Dissect the pipeline
Design and Technical Concern of the Pipeline
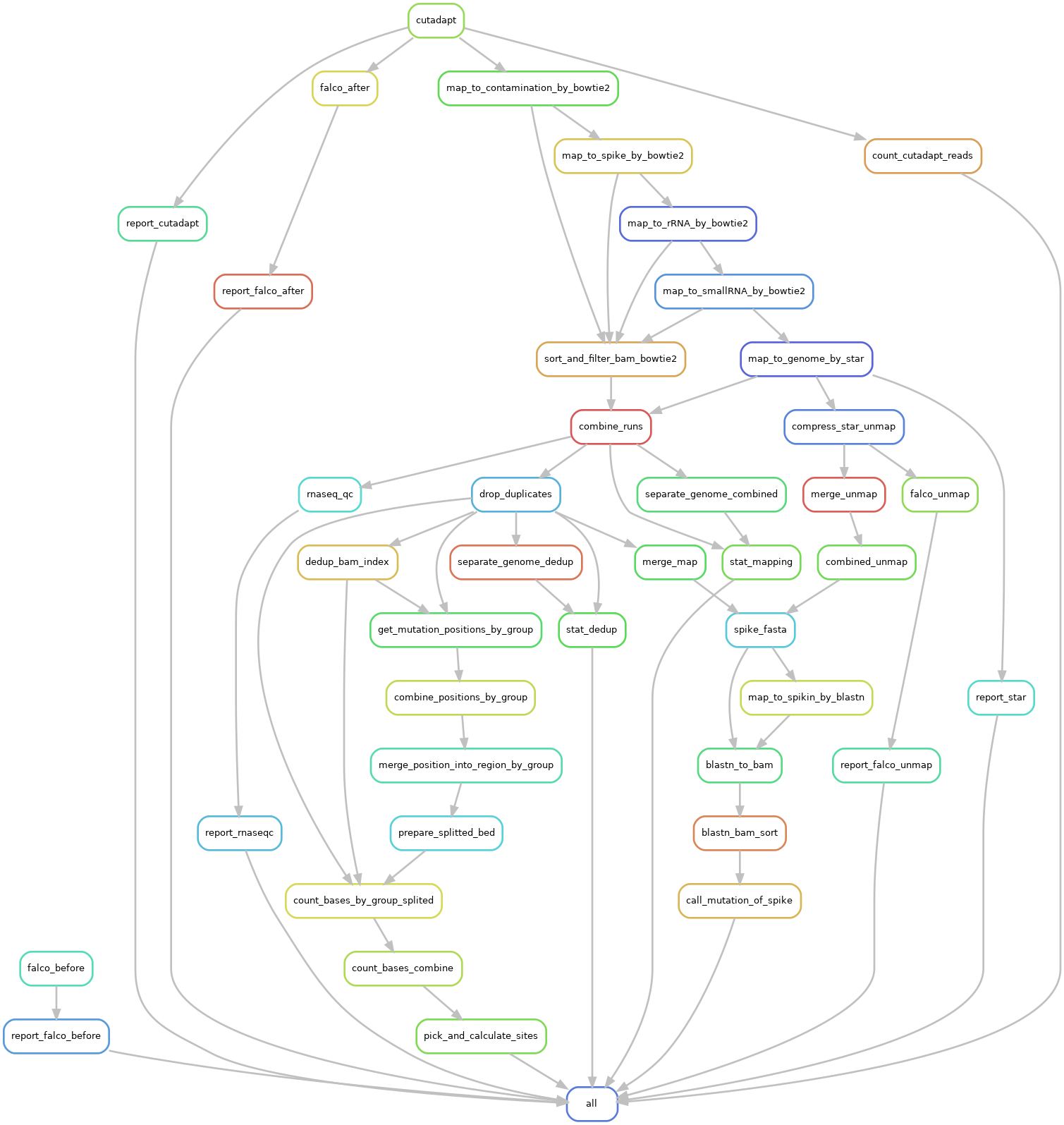
Adapter trimming
concern:
- remove adapter
- remove low quality reads
- remove short reads
- keep UMI for PCR duplicate removal
- random annelling of HIV RT enzyme
- chimera reads
cutadapt -j {threads} \
-U 11 \
--rename='_ ' \
--max-n=0 -e 0.15 -q 20 --nextseq-trim=20 \
-O 6 \
--pair-filter=both \
-a {params.adapter3_r1} -A {params.adapter3_r2} \
-o {output.inter_1} -p {output.inter_2} \
{input} >{output.report1}
cutadapt -j {threads} \
-m 15 \
-u -11 \
-n 5 \
-O 12 \
-g {params.primerF} -a {params.primerR} \
-G {params.primerF} -A {params.primerR} \
--too-short-output={output.short_1} --too-short-paired-output={output.short_2} \
-o {output.trimmed_1} -p {output.trimmed_2} \
{output.inter_1} {output.inter_2} >{output.report2}
Alignment parameters
concern:
- random RT tail
- chimera reads
--alignEndsType Local \
--outFilterMatchNminOverLread 0.66 \
--outFilterMatchNmin 15 \
--outFilterMismatchNmax 5 \
--outFilterMismatchNoverLmax 0.2 \
--outFilterMultimapNmax 50 \
Mutation calling
concern:
- pair end calling
- base quality
- keep minor alternative allele
m6A sites filtering
-
based on hard cutoff
-
based on statistical pval
-
based on modeling